Shiyu Long, Huipeng Yao* , Qi Wu, Guoling LiCollege of Life Science, Sichuan Agriculture University, Ya'an 625014, Sichuan, People’s Republic of China
ABSTRACT
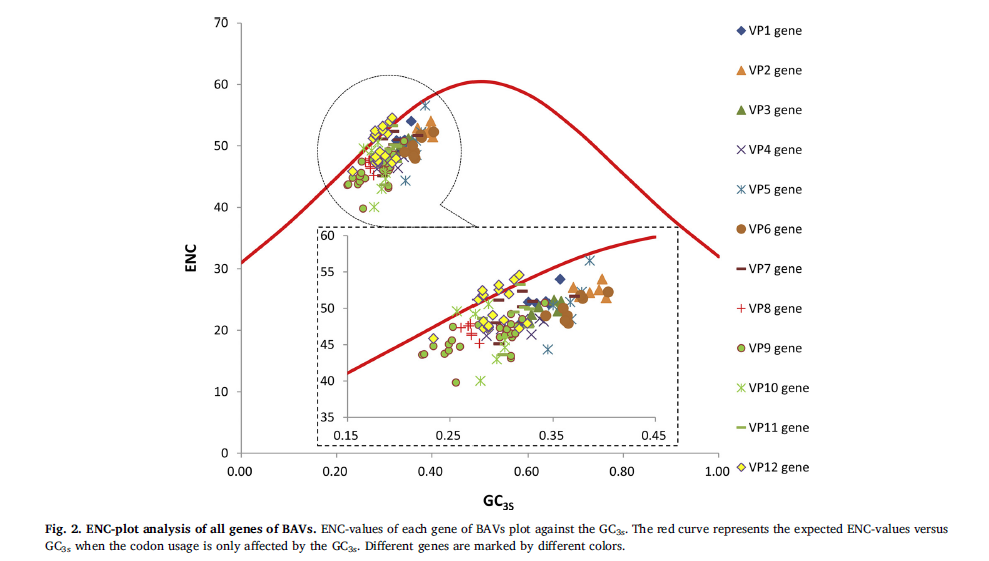
By using DNA Star, CUSP of EMBOSS, Codon W and IBM SPSS Statistics, nucleotide composition and codon usage pattern of 115 genes are researched in 37 BAVs. It shows that the composition of all genes prefers to AU, compared to CG, and for most of genes, the order is A, U, G and C in the virus. The ENC-values of the genes are slightly high which shows the weak codon bias, in which the codon bias of VP9 gene is the highest. The codon usage pattern of 12 different genes is different and related to their composition, their function or their host. For example, VP9 gene encoding viral spike protein in contact with different hosts spread dispersedly and VP1, VP2, VP3 and VP4 genes encoding the capsid protein are concentrated on first quadrant in correspondence analysis. The ENC of VP5 is correlated to GC3s in correlation analysis. The points of VP12 gene are tightly close to the expected curve in the ENC-plot analysis but GC12 is not related to GC3 in neutrality analysis. All analysis indicates the codon usage pattern of 12 genes is influenced by both natural selection and neutral mutation in a different extent in BAV. As a pathogen of viral encephalitis, compositional analysis and codon bias analysis of BAV can provide a theoretical basis for the disease control.
FULL-TEXT: Analysis of compositional bias and codon usage pattern of the coding sequence in Banna virus genome.pdf
Analysis of compositional bias and codon usage pattern of the coding sequence in Banna virus genome.pdf